classical-mds
Classical multi-dimensional scaling (MDS) for dimensionality reduction and low-dimensional embedding. This is also useful for visualizing the similarities of individual items in a 2-dimensional scattered plot.
Header
#include <mathtoolbox/classical-mds.hpp>
Math
Overview
Given a distance (or dissimilarity) matrix of elements
and a target dimensionality , this technique calculates a set of -dimensional coordinates for them:
If the elements are originally defined in an -dimensional space () and Euclidian distance is used for calculating the distance matrix, then this is considered dimensionality reduction (or low-dimensional embedding).
Algorithm
First, calculate the kernel matrix:
where is called the centering matrix and defined as
and is the squared distance matrix.
Then, apply eigenvalue decomposition to :
Finally, pick up the -largest eigenvalues and corresponding eigenvectors , and calculate by
Usage
This technique can be calculated by the following function:
Eigen::MatrixXd ComputeClassicalMds(const Eigen::MatrixXd& D, const unsigned target_dim);
where target_dim is the target dimensionality for embedding.
Example
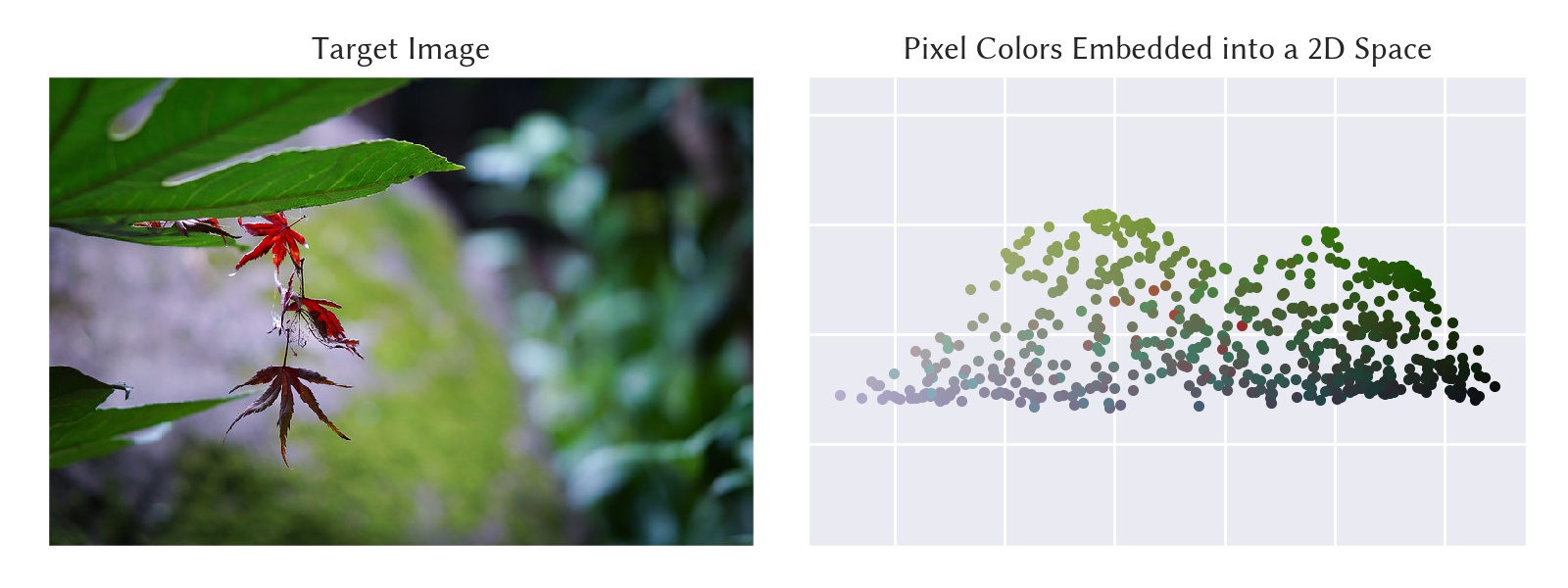
The following is an example of applying the algorithm to the pixel RGB values of a target image and embedding them into a two-dimensional space.
Useful Resources
- Josh Wills, Sameer Agarwal, David Kriegman, and Serge Belongie. 2009. Toward a perceptual space for gloss. ACM Trans. Graph. 28, 4, Article 103 (September 2009), 15 pages. DOI: https://doi.org/10.1145/1559755.1559760